C Fast approximate Bayesian inference
C.1 Epilepsy example
C.1.1 TMB C++ template
// epil.cpp
#include <TMB.hpp>
template <class Type>
Type objective_function<Type>::operator()()
{
DATA_INTEGER(N);
DATA_INTEGER(J);
DATA_INTEGER(K);
DATA_MATRIX(X);
DATA_VECTOR(y);
DATA_MATRIX(E); // Epsilon matrix
PARAMETER_VECTOR(beta);
PARAMETER_VECTOR(epsilon);
PARAMETER_VECTOR(nu);
PARAMETER(l_tau_epsilon);
PARAMETER(l_tau_nu);
Type tau_epsilon = exp(l_tau_epsilon);
Type tau_nu = exp(l_tau_nu);
Type sigma_epsilon = sqrt(1 / tau_epsilon);
Type sigma_nu = sqrt(1 / tau_nu);
vector<Type> eta(X * beta + nu + E * epsilon);
vector<Type> lambda(exp(eta));
Type nll;
nll = Type(0.0);
// Note: dgamma() is parameterised as (shape, scale)
// R-INLA is parameterised as (shape, rate)
nll -= dlgamma(l_tau_epsilon, Type(0.001),
Type(1.0 / 0.001), true);
nll -= dlgamma(l_tau_nu, Type(0.001), Type(1.0 / 0.001), true);
nll -= dnorm(epsilon, Type(0), sigma_epsilon, true).sum();
nll -= dnorm(nu, Type(0), sigma_nu, true).sum();
nll -= dnorm(beta, Type(0), Type(100), true).sum();
nll -= dpois(y, lambda, true).sum();
ADREPORT(tau_epsilon);
ADREPORT(tau_nu);
return(nll);
}C.1.2 Modified TMB C++ template
// epil_modified.cpp
#include <TMB.hpp>
template <class Type>
Type objective_function<Type>::operator()()
{
DATA_INTEGER(N);
DATA_INTEGER(J);
DATA_INTEGER(K);
DATA_MATRIX(X);
DATA_VECTOR(y);
DATA_MATRIX(E); // Epsilon matrix
DATA_IVECTOR(x_starts); // Start index of each subvector of x
DATA_IVECTOR(x_lengths); // Length of each subvector of x
DATA_INTEGER(i); // Index i
PARAMETER(x_i);
PARAMETER_VECTOR(x_minus_i);
vector<Type> x(301);
int k = 0;
for (int j = 0; j < 301; j++) {
if (j + 1 == i) { // +1 because C++ does zero-indexing
x(j) = x_i;
} else {
x(j) = x_minus_i(k);
k++;
}
}
vector<Type> beta = x.segment(x_starts(0), x_lengths(0));
vector<Type> epsilon = x.segment(x_starts(1), x_lengths(1));
vector<Type> nu = x.segment(x_starts(2), x_lengths(2));
PARAMETER(l_tau_epsilon);
PARAMETER(l_tau_nu);
Type tau_epsilon = exp(l_tau_epsilon);
Type tau_nu = exp(l_tau_nu);
Type sigma_epsilon = sqrt(1 / tau_epsilon);
Type sigma_nu = sqrt(1 / tau_nu);
vector<Type> eta(X * beta + nu + E * epsilon);
vector<Type> lambda(exp(eta));
Type nll;
nll = Type(0.0);
// Note: dgamma() is parameterised as (shape, scale)
// R-INLA is parameterised as (shape, rate)
nll -= dlgamma(l_tau_epsilon, Type(0.001),
Type(1.0 / 0.001), true);
nll -= dlgamma(l_tau_nu, Type(0.001), Type(1.0 / 0.001), true);
nll -= dnorm(epsilon, Type(0), sigma_epsilon, true).sum();
nll -= dnorm(nu, Type(0), sigma_nu, true).sum();
nll -= dnorm(beta, Type(0), Type(100), true).sum();
nll -= dpois(y, lambda, true).sum();
ADREPORT(tau_epsilon);
ADREPORT(tau_nu);
return(nll);
}C.1.3 Stan C++ template
// epil.stan
data {
int<lower=0> N; // Number of patients
int<lower=0> J; // Number of clinic visits
int<lower=0> K; // Number of predictors (inc. intercept)
matrix[N * J, K] X; // Design matrix
int<lower=0> y[N * J]; // Outcome variable
matrix[N * J, N] E; // Epsilon matrix
}
parameters {
vector[K] beta; // Vector of coefficients
vector[N] epsilon; // Patient specific errors
vector[N * J] nu; // Patient-visit errors
real<lower=0> tau_epsilon; // Precision of epsilon
real<lower=0> tau_nu; // Precision of nu
}
transformed parameters {
vector[N * J] eta = X * beta + nu + E * epsilon;
}
model {
beta ~ normal(0, 100);
tau_epsilon ~ gamma(0.001, 0.001);
tau_nu ~ gamma(0.001, 0.001);
epsilon ~ normal(0, sqrt(1 / tau_epsilon));
nu ~ normal(0, sqrt(1 / tau_nu));
y ~ poisson_log(eta);
}C.1.4 NUTS convergence and suitability
C.1.4.1 tmbstan
![Traceplots for the tmbstan parameters with the lowest ESS and highest potential scale reduction factor. These were l_tau_nu (an \(\text{ESS}\) of 377) and beta[3] (an \(\hat R\) of 1.006).](figures/naomi-aghq/tmbstan-epil.png)
Figure C.1: Traceplots for the tmbstan parameters with the lowest ESS and highest potential scale reduction factor. These were l_tau_nu (an \(\text{ESS}\) of 377) and beta[3] (an \(\hat R\) of 1.006).
C.1.4.2 rstan
![Traceplots for the rstan parameters with the lowest ESS and highest potential scale reduction factor. These were tau_nu (an \(\text{ESS}\) of 437) and tau_nu (an \(\hat R\) of 1.009). Rather than plotting the traceplot for tau_nu twice, the parameter epsilon[18] is included, which had the second highest \(\hat R\) of 1.008.](figures/naomi-aghq/stan-epil.png)
Figure C.2: Traceplots for the rstan parameters with the lowest ESS and highest potential scale reduction factor. These were tau_nu (an \(\text{ESS}\) of 437) and tau_nu (an \(\hat R\) of 1.009). Rather than plotting the traceplot for tau_nu twice, the parameter epsilon[18] is included, which had the second highest \(\hat R\) of 1.008.
C.2 Loa loa example
C.2.1 NUTS convergence and suitability
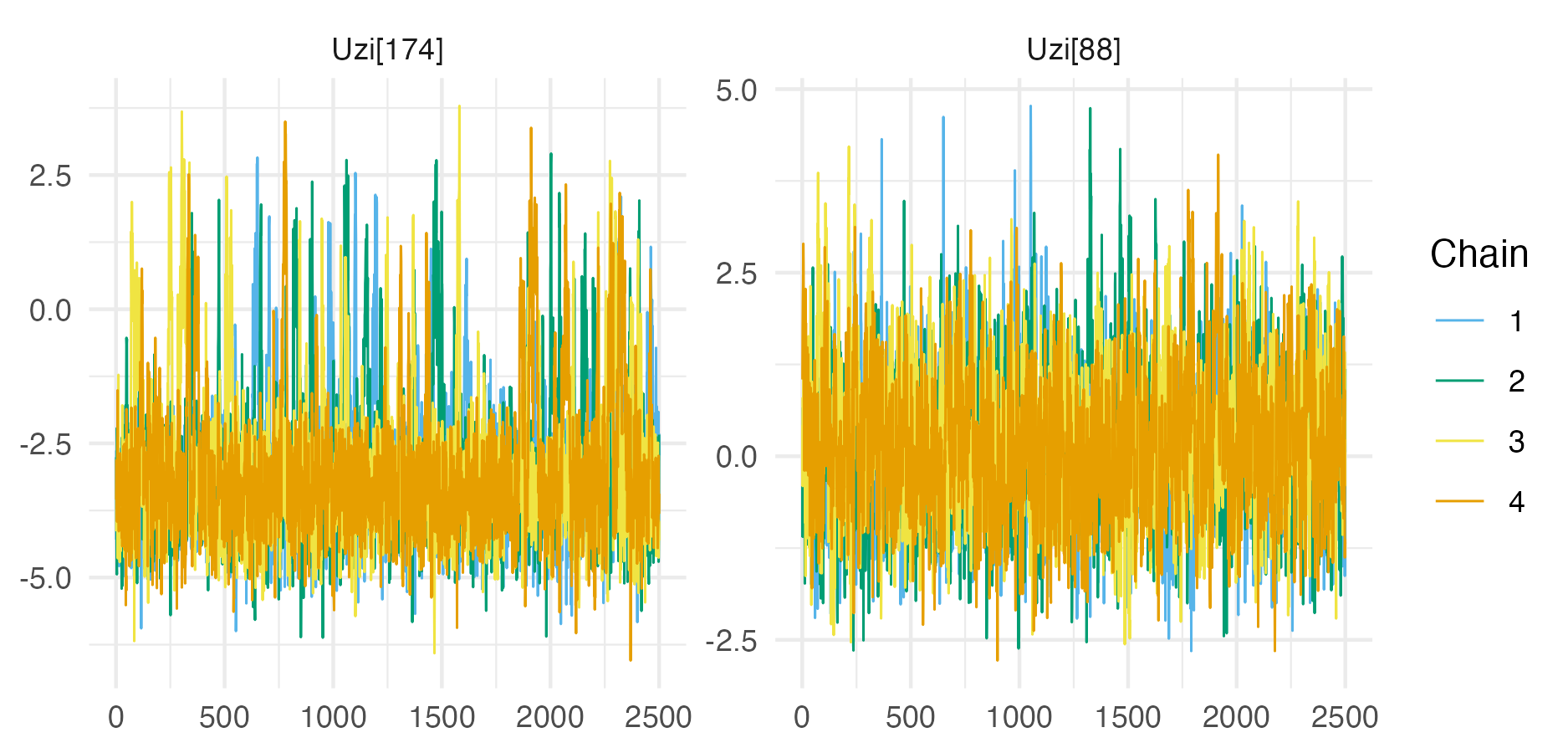
Figure C.3: Traceplots for the parameters with the lowest ESS and highest potential scale reduction factor for the Loa loa ELGM example.
C.2.2 Inference comparison
![Relative difference between the Gaussian and Laplace marginal posterior means and standard deviations to NUTS results at each \(u(s_i), v(s_i): i \in [190]\). Absolute differences are in Figure 6.14.](figures/naomi-aghq/loa-loa-mean-sd-pct.png)
Figure C.4: Relative difference between the Gaussian and Laplace marginal posterior means and standard deviations to NUTS results at each \(u(s_i), v(s_i): i \in [190]\). Absolute differences are in Figure 6.14.
C.3 AGHQ with Laplace marginals algorithm
This section provides the INLA-like algorithm for AGHQ with Laplace marginals used in this thesis.
The algorithm for AGHQ with Gaussian marginals used in this thesis is as given in Stringer, Brown, and Stafford (2022), and implemented in the aghq package.
Calculate the mode, Hessian at the mode, lower Cholesky, and Laplace approximation \[\begin{align} \hat{\boldsymbol{\mathbf{\theta}}} &= \arg \max_{\boldsymbol{\mathbf{\theta}}} {\tilde p_\texttt{LA}(\boldsymbol{\mathbf{\theta}}, \mathbf{y})}, \\ \hat{\mathbf{H}} &= - \frac{\partial^2}{\partial \boldsymbol{\mathbf{\theta}} \partial \boldsymbol{\mathbf{\theta}}^\top} \log \tilde p_\texttt{LA}(\boldsymbol{\mathbf{\theta}}, \mathbf{y}) \rvert_{\boldsymbol{\mathbf{\theta}} = \hat{\boldsymbol{\mathbf{\theta}}}}, \\ \hat{\mathbf{H}}^{-1} &= \hat{\mathbf{L}} \hat{\mathbf{L}}^\top, \\ \tilde p_\texttt{LA}(\boldsymbol{\mathbf{\theta}}, \mathbf{y}) &= \frac{p(\mathbf{y}, \mathbf{x}, \boldsymbol{\mathbf{\theta}})}{\tilde p_\texttt{G}(\mathbf{x} \, | \, \boldsymbol{\mathbf{\theta}}, \mathbf{y})} \Big\rvert_{\mathbf{x} = \hat{\mathbf{x}}(\boldsymbol{\mathbf{\theta}})}, \end{align}\] where \(\tilde p_\texttt{G}(\mathbf{x} \, | \, \boldsymbol{\mathbf{\theta}}, \mathbf{y}) = \mathcal{N}(\mathbf{x} \, | \, \hat{\mathbf{x}}(\boldsymbol{\mathbf{\theta}}), \hat{\mathbf{H}}(\boldsymbol{\mathbf{\theta}})^{-1})\) is a Gaussian approximation to \(p(\mathbf{x} \, | \, \boldsymbol{\mathbf{\theta}}, \mathbf{y})\) with mode and precision matrix given by \[\begin{align} \hat{\mathbf{x}}(\boldsymbol{\mathbf{\theta}}) &= \arg \max_\mathbf{x} \log p(\mathbf{y}, \mathbf{x}, \boldsymbol{\mathbf{\theta}}), \\ \hat{\mathbf{H}}(\boldsymbol{\mathbf{\theta}}) &= - \frac{\partial^2}{\partial \mathbf{x} \partial \mathbf{x}^\top} \log p(\mathbf{y}, \mathbf{x}, \boldsymbol{\mathbf{\theta}}) \rvert_{\mathbf{x} = \hat{\mathbf{x}}(\boldsymbol{\mathbf{\theta}})}. \end{align}\]
Generate a set of nodes \(\mathbf{u} \in \mathcal{Q}(m, k)\) and weights \(\omega: \mathbf{u} \to \mathbb{R}\) from a Gauss-Hermite quadrature rule with \(k\) nodes per dimension. Adapt these nodes based on the mode and lower Cholesky via \(\boldsymbol{\mathbf{\theta}}(\mathbf{u}) = \hat{\boldsymbol{\mathbf{\theta}}} + \mathbf{L} \mathbf{u}\). Use this quadrature rule to calculate the normalising constant \(\tilde p_{\texttt{AQ}}(\mathbf{y})\) as follows \[\begin{equation} \tilde p_{\texttt{AQ}}(\mathbf{y}) = \sum_{\mathbf{u} \in \mathcal{Q}(m, k)} \tilde p_\texttt{LA}(\boldsymbol{\mathbf{\theta}}(\mathbf{u}), \mathbf{y}) \omega(\mathbf{u}). \tag{C.1} \end{equation}\]
For \(i \in [N]\) generate \(l\) nodes \(x_i(\mathbf{v})\) via a Gauss-Hermite quadrature rule \(\mathbf{v} \in \mathcal{Q}(1, l)\) adapted based on the mode \(\hat{\mathbf{x}}(\boldsymbol{\mathbf{\theta}})_i\) and standard deviation \(\sqrt{\text{diag}[\hat{\mathbf{H}}(\boldsymbol{\mathbf{\theta}})^{-1}]_i}\) of the Gaussian marginal. A value of \(l \geq 4\) is recommended to enable B-spline interpolation. For \(x_i \in \{ x_i(\mathbf{v}) \}_{\mathbf{v} \in \mathcal{Q}(1, l)}\) and \(\boldsymbol{\mathbf{\theta}} \in \{ \boldsymbol{\mathbf{\theta}}(\mathbf{u}) \}_{\mathbf{u} \in \mathcal{Q}(m, k)}\) calculate the modes and Hessians \[\begin{align} \hat{\mathbf{x}}_{-i}(x_i, \boldsymbol{\mathbf{\theta}}) &= \arg \max_{\mathbf{x}_{-i}} \log p(\mathbf{y}, x_i, \mathbf{x}_{-i}, \boldsymbol{\mathbf{\theta}}), \\ \hat{\mathbf{H}}_{-i, -i}(x_i, \boldsymbol{\mathbf{\theta}}) &= - \frac{\partial^2}{\partial \mathbf{x}_{-i} \partial \mathbf{x}_{-i}^\top} \log p(\mathbf{y}, x_i, \mathbf{x}_{-i}, \boldsymbol{\mathbf{\theta}}) \rvert_{\mathbf{x}_{-i} = \hat{\mathbf{x}}_{-i}(x_i, \boldsymbol{\mathbf{\theta}})}, \end{align}\] where optimisation to obtain \(\hat{\mathbf{x}}_{-i}(x_i, \boldsymbol{\mathbf{\theta}})\) can be initialised at \(\hat{\mathbf{x}}(\boldsymbol{\mathbf{\theta}})_{-i}\).
For \(x_i \in \{ x_i(\mathbf{v}) \}_{\mathbf{v} \in \mathcal{Q}(1, l)}\) calculate \[\begin{equation} p_\texttt{AQ}(x_i \, | \, \mathbf{y}) = \frac{\tilde p_\texttt{LA}(x_i, \mathbf{y})}{\tilde p_{\texttt{AQ}}(\mathbf{y})}, \tag{C.2} \end{equation}\] where \[\begin{equation} \tilde p_\texttt{LA}(x_i, \mathbf{y}) = \sum_{\mathbf{u} \in \mathcal{Q}(m, k)} \tilde p_\texttt{LA}(x_i, \boldsymbol{\mathbf{\theta}}(\mathbf{u}), \mathbf{y}) \omega(\mathbf{u}). \end{equation}\] and \[\begin{equation} \tilde p_\texttt{LA}(x_i, \boldsymbol{\mathbf{\theta}}, \mathbf{y}) = \frac{p(x_i, \mathbf{x}_{-i}, \boldsymbol{\mathbf{\theta}}, \mathbf{y})}{\tilde p_\texttt{G}(\mathbf{x}_{-i} \, | \, x_i, \boldsymbol{\mathbf{\theta}}, \mathbf{y})} \Big\rvert_{\mathbf{x}_{-i} = \hat{\mathbf{x}}_{-i}(x_i, \boldsymbol{\mathbf{\theta}})}. \end{equation}\] Equation (C.2) can be calculated using the estimate of the evidence given in Equation (C.1), but it is more numerically accurate, and requires little extra computation, to use the estimate \[\begin{equation} \tilde p_{\texttt{AQ}}(\mathbf{y}) = \sum_{\mathbf{v} \in \mathcal{Q}(1, l)} \tilde p_\texttt{LA}(x_i(\mathbf{v}), \mathbf{y}) \omega(\mathbf{v}) \end{equation}\]
Given \(\{x_i(\mathbf{v}), \tilde p_\texttt{AQ}(x_i(\mathbf{v}) \, | \, \mathbf{y})\}_{\mathbf{v} \in \mathcal{Q}(1, l)}\) create a spline interpolant to each posterior marginal on the log-scale. Samples, and thereby relevant posterior marginal summaries, may be obtained using inverse transform sampling.
C.4 Simplified Naomi model description
This section describes the simplified version of the Naomi model (Eaton et al. 2021) in more detail. The concise \(i\) indexing used in Section 6.3 is replaced by a more complete \(x, s, a\) indexing. There are four sections:
- Section C.4.1 gives the process specifications, giving the terms in each structured additive predictor, along with their distributions.
- Section C.4.2 gives additional details about the likelihood terms not provided in Section 6.3.
- Section C.4.3 gives identifiability constraints used in circumstances where incomplete data is available for the country.
- Section C.4.4 provides details of the
TMBimplementation.
C.4.1 Process specification
| Model component | Latent field | Hyperparameter | |
|---|---|---|---|
| Section C.4.1.1 | HIV prevalence | \(22 + 5n\) | 9 |
| Section C.4.1.2 | ART coverage | \(25 + 5n\) | 9 |
| Section C.4.1.3 | HIV incidence rate | \(2 + n\) | 3 |
| Section C.4.1.4 | ANC testing | \(2 + 2n\) | 2 |
| Section C.4.1.5 | ART attendance | \(n\) | 1 |
| Total | \(51 + 14n\) | 24 |
C.4.1.1 HIV prevalence
HIV prevalence \(\rho_{x, s, a} \in [0, 1]\) was modelled on the logit scale using the structured additive predictor \[\begin{equation} \text{logit}(\rho_{x, s, a}) = \beta^\rho_0 + \beta_{S}^{\rho, s = \text{M}} + \mathbf{u}^\rho_a + \mathbf{u}_a^{\rho, s = \text{M}} + \mathbf{u}^\rho_x + \mathbf{u}_x^{\rho, s = \text{M}} + \mathbf{u}_x^{\rho, a < 15} + \boldsymbol{\mathbf{\eta}}^\rho_{R_x, s, a}. \tag{C.3} \end{equation}\] Table C.2 provides a description of the terms included in Equation (C.3). Independent half-normal prior distributions were chosen for the five standard deviation terms \[\begin{equation} \{\sigma_A^\rho, \sigma_{AS}^\rho, \sigma_X^\rho, \sigma_{XS}^\rho, \sigma_{XA}^\rho\} \sim \mathcal{N}^{+}(0, 2.5), \end{equation}\] independent uniform prior distributions for the two AR1 correlation parameters \[\begin{equation} \{\phi_A^\rho, \phi_{AS}^\rho\} \sim \mathcal{U}(-1, 1), \end{equation}\] and independent beta prior distributions for the two BYM2 proportion parameters \[\begin{equation} \{\phi_X^\rho, \phi_{XS}^\rho\} \sim \text{Beta}(0.5, 0.5). \end{equation}\]
| Term | Distribution | Description |
|---|---|---|
| \(\beta^\rho_0\) | \(\mathcal{N}(0, 5)\) | Intercept |
| \(\beta_{s}^{\rho, s = \text{M}}\) | \(\mathcal{N}(0, 5)\) | The difference in logit prevalence for men compared to women |
| \(\mathbf{u}^\rho_a\) | \(\text{AR}1(\sigma_A^\rho, \phi_A^\rho)\) | Age random effects for women |
| \(\mathbf{u}_a^{\rho, s = \text{M}}\) | \(\text{AR}1(\sigma_{AS}^\rho, \phi_{AS}^\rho)\) | Age random effects for the difference in logit prevalence for men compared to women age \(a\) |
| \(\mathbf{u}^\rho_x\) | \(\text{BYM}2(\sigma_X^\rho, \phi_X^\rho)\) | Spatial random effects for women |
| \(\mathbf{u}_x^{\rho, s = \text{M}}\) | \(\text{BYM}2(\sigma_{XS}^\rho, \phi_{XS}^\rho)\) | Spatial random effects for the difference in logit prevalence for men compared to women in district \(x\) |
| \(\mathbf{u}_x^{\rho, a < 15}\) | \(\text{ICAR}(\sigma_{XA}^\rho)\) | Spatial random effects for the difference in logit paediatric prevalence to adult women prevalence in district \(x\) |
| \(\boldsymbol{\mathbf{\eta}}^\rho_{R_x, s, a}\) | \(-\) | Fixed offsets specifying assumed odds ratios for prevalence outside the age ranges for which data were available. Calculated from Spectrum model (Stover et al. 2019) outputs for region \(R_x\) |
C.4.1.2 ART coverage
ART coverage \(\alpha_{x, s, a} \in [0, 1]\) was modelled on the logit scale using the structured additive predictor \[\begin{equation} \text{logit}(\alpha_{x, s, a}) = \beta^\alpha_0 + \beta_{S}^{\alpha, s = \text{M}} + \mathbf{u}^\alpha_a + \mathbf{u}_a^{\alpha, s = \text{M}} + \mathbf{u}^\alpha_x + \mathbf{u}_x^{\alpha, s = \text{M}} + \mathbf{u}_x^{\alpha, a < 15} + \boldsymbol{\mathbf{\eta}}^\alpha_{R_x, s, a} \end{equation}\] with terms and prior distributions analogous to the HIV prevalence process model in Section C.4.1.1 above.
C.4.1.3 HIV incidence rate
HIV incidence rate \(\lambda_{x, s, a} > 0\) was modelled on the log scale using the structured additive predictor \[\begin{equation} \log(\lambda_{x, s, a}) = \beta_0^\lambda + \beta_S^{\lambda, s = \text{M}} + \log(\rho_{x}^{\text{15-49}}) + \log(1 - \omega \cdot \alpha_{x}^{\text{15-49}}) + \mathbf{u}_x^\lambda + \boldsymbol{\mathbf{\eta}}_{R_x, s, a}^\lambda. \tag{C.4} \end{equation}\] Table C.3 provides a description of the terms included in Equation (C.4).
| Term | Distribution | Description |
|---|---|---|
| \(\beta^\lambda_0\) | \(\mathcal{N}(0, 5)\) | Intercept term proportional to the average HIV transmission rate for untreated HIV positive adults |
| \(\beta_S^{\lambda, s = \text{M}}\) | \(\mathcal{N}(0, 5)\) | The log incidence rate ratio for men compared to women |
| \(\rho_{x}^{\text{15-49}}\) | \(-\) | The HIV prevalence among adults 15-49 in district \(x\) calculated by aggregating age-specific HIV prevalences |
| \(\alpha_{x}^{\text{15-49}}\) | \(-\) | The ART coverage among adults 15-49 in district \(x\) calculated by aggregating age-specific ART coverages |
| \(\omega = 0.7\) | \(-\) | Average reduction in HIV transmission rate per increase in population ART coverage fixed based on inputs to the Estimation and Projection Package (EPP) model |
| \(\mathbf{u}_x^\lambda\) | \(\mathcal{N}(0, \sigma^\lambda)\) | IID spatial random effects with \(\sigma^\lambda \sim \mathcal{N}^+(0, 1)\) |
| \(\boldsymbol{\mathbf{\eta}}^\lambda_{R_x, s, a}\) | \(-\) | Fixed log incidence rate ratios by sex and age group calculated from Spectrum model outputs for region \(R_x\) |
The proportion recently infected among HIV positive persons \(\kappa_{x, s, a} \in [0, 1]\) was modelled as \[\begin{equation} \kappa_{x, s, a} = 1 - \exp \left(- \lambda_{x, s, a} \cdot \frac{1 - \rho_{x, s, a}}{\rho_{x, s, a}} \cdot (\Omega_T - \beta_T ) - \beta_T \right), \end{equation}\] where \(\Omega_T \sim \mathcal{N}(\Omega_{T_0}, \sigma^{\Omega_T})\) is the mean duration of recent infection, and \(\beta_T \sim \mathcal{N}^{+}(\beta_{T_0}, \sigma^{\beta_T})\) is the false recent ratio. The prior distribution for \(\Omega_T\) was informed by the characteristics of the recent infection testing algorithm. For PHIA surveys this was \(\Omega_{T_0} = 130 \text{ days}\) and \(\sigma^{\Omega_T} = 6.12 \text{ days}\). For PHIA surveys there was assumed to be no false recency, such that \(\beta_{T_0} = 0.0\), \(\sigma^{\beta_T} = 0.0\), and \(\beta_T = 0\).
C.4.1.4 ANC testing
HIV prevalence \(\rho_{x, a}^\text{ANC}\) and ART coverage \(\alpha_{x, a}^\text{ANC}\) among pregnant women were modelled as being offset on the logit scale from the corresponding district-age indicators \(\rho_{x, F, a}\) and \(\alpha_{x, F, a}\) according to \[\begin{align} \text{logit}(\rho_{x, a}^{\text{ANC}}) &= \text{logit}(\rho_{x, F, a}) + \beta^{\rho^{\text{ANC}}} + \mathbf{u}_x^{\rho^{\text{ANC}}} + \boldsymbol{\mathbf{\eta}}_{R_x, a}^{\rho^{\text{ANC}}}, \tag{C.5} \\ \text{logit}(\alpha_{x, a}^{\text{ANC}}) &= \text{logit}(\alpha_{x, F, a}) + \beta^{\alpha^{\text{ANC}}} + \mathbf{u}_x^{\alpha^{\text{ANC}}} + \boldsymbol{\mathbf{\eta}}_{R_x, a}^{\alpha^{\text{ANC}}} \tag{C.6}. \end{align}\] Table C.4 provides a description of the terms included in Equation (C.5) and Equation (C.6).
| Term | Distribution | Description |
|---|---|---|
| \(\beta^{\theta^{\text{ANC}}}\) | \(\mathcal{N}(0, 5)\) | Intercept giving the average difference between population and ANC outcomes |
| \(\mathbf{u}_x^{\theta^{\text{ANC}}}\) | \(\mathcal{N}(0, \sigma_X^{\theta^{\text{ANC}}})\) | IID district random effects with \(\sigma_X^{\theta^{\text{ANC}}} \sim \mathcal{N}^+(0, 1)\) |
| \(\boldsymbol{\mathbf{\eta}}_{R_x, a}^{\theta^{\text{ANC}}}\) | \(-\) | Offsets for the log fertility rate ratios for HIV positive women compared to HIV negative women and for women on ART to HIV positive women not on ART, calculated from Spectrum model outputs for region \(R_x\) |
In the full Naomi model, for adult women 15-49 the number of ANC clients \(\Psi_{x, a} > 0\) were modelled as \[\begin{equation} \log (\Psi_{x, a}) = \log (N_{x, \text{F}, a}) + \psi_{R_x, a} + \beta^\psi + \mathbf{u}_x^\psi, \end{equation}\] where \(N_{x, \text{F}, a}\) are the female population sizes, \(\psi_{R_x, a}\) are fixed age-sex fertility ratios in Spectrum region \(R_x\), \(\beta^\psi\) are log rate ratios for the number of ANC clients relative to the predicted fertility, and \(\mathbf{u}_x^\psi \sim \mathcal{N}(0, \sigma^\psi)\) are district random effects. Here these terms are fixed to \(\beta^\psi = 0\) and \(\mathbf{u}_x^\psi = \mathbf{0}\) such that \(\Psi_{x, a}\) are simply constants.
C.4.1.5 ART attendance
Let \(\gamma_{x, x'} \in [0, 1]\) be the probability that a person on ART residing in district \(x\) receives ART in district \(x'\). Assume that \(\gamma_{x, x'} = 0\) for \(x \notin \{x, \text{ne}(x)\}\) such that individuals seek treatment only in their residing district or its neighbours \(\text{ne}(x) = \{x': x' \sim x\}\), where \(\sim\) is an adjacency relation, and \(\sum_{x' \in \{x, \text{ne}(x)\}} \gamma_{x, x'} = 1\).
The probabilities \(\gamma_{x, x'}\) for \(x \sim x'\) were modelled using multinomial logistic regression model, based on the log-odds ratios \[\begin{equation} \tilde \gamma_{x, x'} = \log \left( \frac{\gamma_{x, x'}}{1 - \gamma_{x, x'}} \right) = \tilde \gamma_0 + \mathbf{u}_x^{\tilde \gamma}. \tag{C.7} \end{equation}\] Table C.5 provides a description of the terms included in Equation (C.7). Fixing \(\tilde \gamma_{x, x} = 0\) then the multinomial probabilities may be recovered using the softmax \[\begin{equation} \gamma_{x, x'} = \frac{\exp(\tilde \gamma_{x, x'})}{\sum_{x^\star \in \{x, \text{ne}(x)\}} \exp(\tilde \gamma_{x, x^\star})}. \end{equation}\]
| Term | Distribution | Description |
|---|---|---|
| \(\tilde \gamma_0\) | \(-\) | Fixed intercept \(\tilde \gamma_0 = -4\). Implies a prior mean on \(\gamma_{x, x'}\) of 1.8%, such that a-priori \((100 - 1.8 \times \text{ne}(x))\%\) of ART clients in district \(x\) obtain treatment in their home district |
| \(\mathbf{u}_x^{\tilde \gamma}\) | \(\mathcal{N}(0, \sigma_X^{\tilde \gamma})\) | District random effects, with \(\sigma_X^{\tilde \gamma} \sim \mathcal{N}^+(0, 2.5)\) |
C.4.2 Additional likelihood specification
Though Section 6.3 provides a complete description of Naomi’s likelihood specification, any additional useful details are provided here.
C.4.2.1 Household survey data
The generalised binomial \(y \sim \text{xBin}(m, p)\) is defined for \(y, m \in \mathbb{R}^+\) with \(y \leq m\) such that \[\begin{align} \log p(y) = &\log \Gamma(m + 1) - \log \Gamma(y + 1) \\ &- \log \Gamma(m - y + 1) + y \log p + (m - y) \log(1 - p), \end{align}\] where the gamma function \(\Gamma\) is such that \(\forall n \in \mathbb{N}\), \(\Gamma(n) = (n - 1)!\).
C.4.3 Identifiability constraints
If data are missing, some parameters are fixed to default values to help with identifiability. In particular:
- If survey data on HIV prevalence or ART coverage by age and sex are not available then \(\mathbf{u}_a^\theta = 0\) and \(\mathbf{u}_{a, s = \text{M}}^\theta = 0\). In this case, the average age-sex pattern from the Spectrum is used. For the Malawi case-study (Section 6.5), HIV prevalence and ART coverage data are not available for those aged 65+. As a result, there are \(|\{\text{0-4}, \ldots, \text{50-54}\}| = 13\) age groups included for the age random effects.
- If no ART data, either survey or ART programme, are available but data on ART coverage among ANC clients are available, the level of ART coverage is not identifiable, but spatial variation is identifiable. In this instance, overall ART coverage is determined by the Spectrum offset, and only area random effects are estimated such that \[\begin{equation} \text{logit} \left(\alpha_{x, s, a} \right) = \mathbf{u}_x^\alpha + \boldsymbol{\mathbf{\eta}}_{R_x, s, a}^\alpha. \end{equation}\]
- If survey data on recent HIV infection are not included in the model, then \(\beta_0^\lambda = \beta_S^{\lambda, s = \text{M}} = 0\) and \(\mathbf{u}_x^\lambda = \mathbf{0}\). The sex ratio for HIV incidence is determined by the sex incidence rate ratio from Spectrum, and the incidence rate in all districts is modelled assuming the same average HIV transmission rate for untreated adults, but varies according to district-level estimates of HIV prevalence and ART coverage.
C.4.4 Implementation
The TMB C++ code for the negative log-posterior of the simplified Naomi model is available from https://github.com/athowes/naomi-aghq.
For ease of understanding, Table C.6 provides correspondence between the mathematical notation used in Section C.4 and the variable names used in the TMB code, for all hyperparameters and latent field parameters.
For further reference on the TMB software see Kristensen (2021).
| Variable name | Notation | Type | Domain | \(\rho\) | \(\alpha\) | \(\lambda\) |
|---|---|---|---|---|---|---|
logit_phi_rho_x |
\(\text{logit}(\phi_X^\rho)\) | Hyper | \(\mathbb{R}\) | Yes | ||
log_sigma_rho_x |
\(\log(\sigma_X^\rho)\) | Hyper | \(\mathbb{R}\) | Yes | ||
logit_phi_rho_xs |
\(\text{logit}(\phi_{XS}^\rho)\) | Hyper | \(\mathbb{R}\) | Yes | ||
log_sigma_rho_xs |
\(\log(\sigma_{XS}^\rho)\) | Hyper | \(\mathbb{R}\) | Yes | ||
logit_phi_rho_a |
\(\text{logit}(\phi_A^\rho)\) | Hyper | \(\mathbb{R}\) | Yes | ||
log_sigma_rho_a |
\(\log(\sigma_A^\rho)\) | Hyper | \(\mathbb{R}\) | Yes | ||
logit_phi_rho_as |
\(\text{logit}(\phi_{AS}^\rho)\) | Hyper | \(\mathbb{R}\) | Yes | ||
log_sigma_rho_as |
\(\log(\sigma_{AS}^\rho)\) | Hyper | \(\mathbb{R}\) | Yes | ||
log_sigma_rho_xa |
\(\log(\sigma_{XA}^\rho)\) | Hyper | \(\mathbb{R}\) | Yes | ||
logit_phi_alpha_x |
\(\text{logit}(\phi_X^\alpha)\) | Hyper | \(\mathbb{R}\) | Yes | ||
log_sigma_alpha_x |
\(\log(\sigma_X^\alpha)\) | Hyper | \(\mathbb{R}\) | Yes | ||
logit_phi_alpha_xs |
\(\text{logit}(\phi_{XS}^\alpha)\) | Hyper | \(\mathbb{R}\) | Yes | ||
log_sigma_alpha_xs |
\(\log(\sigma_{XS}^\alpha)\) | Hyper | \(\mathbb{R}\) | Yes | ||
logit_phi_alpha_a |
\(\text{logit}(\phi_A^\alpha)\) | Hyper | \(\mathbb{R}\) | Yes | ||
log_sigma_alpha_a |
\(\log(\sigma_A^\alpha)\) | Hyper | \(\mathbb{R}\) | Yes | ||
logit_phi_alpha_as |
\(\text{logit}(\phi_{AS}^\alpha)\) | Hyper | \(\mathbb{R}\) | Yes | ||
log_sigma_alpha_as |
\(\log(\sigma_{AS}^\alpha)\) | Hyper | \(\mathbb{R}\) | Yes | ||
log_sigma_alpha_xa |
\(\log(\sigma_{XA}^\alpha)\) | Hyper | \(\mathbb{R}\) | Yes | ||
OmegaT_raw |
\(\Omega_T\) | Hyper | \(\mathbb{R}\) | Yes | ||
log_betaT |
\(\log(\beta_T)\) | Hyper | \(\mathbb{R}\) | Yes | ||
log_sigma_lambda_x |
\(\log(\sigma^\lambda)\) | Hyper | \(\mathbb{R}\) | Yes | ||
log_sigma_ancrho_x |
\(\log(\sigma_X^{\rho^{\text{ANC}}})\) | Hyper | \(\mathbb{R}\) | Yes | ||
log_sigma_ancalpha_x |
\(\log(\sigma_X^{\alpha^{\text{ANC}}})\) | Hyper | \(\mathbb{R}\) | Yes | ||
log_sigma_or_gamma |
\(\log(\sigma_X^{\tilde \gamma})\) | Hyper | \(\mathbb{R}\) | |||
beta_rho |
\((\beta^\rho_0, \beta_{s}^{\rho, s = \text{M}})\) | Latent | \(\mathbb{R}^2\) | Yes | ||
beta_alpha |
\((\beta^\alpha_0, \beta_{S}^{\alpha, s = \text{M}})\) | Latent | \(\mathbb{R}^2\) | Yes | ||
beta_lambda |
\((\beta_0^\lambda, \beta_S^{\lambda, s = \text{M}})\) | Latent | \(\mathbb{R}^2\) | Yes | ||
beta_anc_rho |
\(\beta^{\rho^{\text{ANC}}}\) | Latent | \(\mathbb{R}\) | Yes | ||
beta_anc_alpha |
\(\beta^{\alpha^{\text{ANC}}}\) | Latent | \(\mathbb{R}\) | Yes | ||
u_rho_x |
\(\mathbf{w}^\rho_x\) | Latent | \(\mathbb{R}^{n}\) | Yes | ||
us_rho_x |
\(\mathbf{v}^\rho_x\) | Latent | \(\mathbb{R}^{n}\) | Yes | ||
u_rho_xs |
\(\mathbf{w}_x^{\rho, s = \text{M}}\) | Latent | \(\mathbb{R}^{n}\) | Yes | ||
us_rho_xs |
\(\mathbf{v}_x^{\rho, s = \text{M}}\) | Latent | \(\mathbb{R}^{n}\) | Yes | ||
u_rho_a |
\(\mathbf{u}^\rho_a\) | Latent | \(\mathbb{R}^{10}\) | Yes | ||
u_rho_as |
\(\mathbf{u}_a^{\rho, s = \text{M}}\) | Latent | \(\mathbb{R}^{10}\) | Yes | ||
u_rho_xa |
\(\mathbf{u}_x^{\rho, a < 15}\) | Latent | \(\mathbb{R}^{n}\) | Yes | ||
u_alpha_x |
\(\mathbf{w}^\alpha_x\) | Latent | \(\mathbb{R}^{n}\) | Yes | ||
us_alpha_x |
\(\mathbf{v}^\alpha_x\) | Latent | \(\mathbb{R}^{n}\) | Yes | ||
u_alpha_xs |
\(\mathbf{w}_x^{\alpha, s = \text{M}}\) | Latent | \(\mathbb{R}^{n}\) | Yes | ||
us_alpha_xs |
\(\mathbf{v}_x^{\alpha, s = \text{M}}\) | Latent | \(\mathbb{R}^{n}\) | Yes | ||
u_alpha_a |
\(\mathbf{u}^\alpha_a\) | Latent | \(\mathbb{R}^{13}\) | Yes | ||
u_alpha_as |
\(\mathbf{u}_a^{\alpha, s = \text{M}}\) | Latent | \(\mathbb{R}^{10}\) | Yes | ||
u_alpha_xa |
\(\mathbf{u}_x^{\alpha, a < 15}\) | Latent | \(\mathbb{R}^{n}\) | Yes | ||
ui_lambda_x |
\(\mathbf{u}_x^\lambda\) | Latent | \(\mathbb{R}^{n}\) | Yes | ||
ui_anc_rho_x |
\(\mathbf{u}_x^{\rho^{\text{ANC}}}\) | Latent | \(\mathbb{R}^{n}\) | Yes | ||
ui_anc_alpha_x |
\(\mathbf{u}_x^{\alpha^{\text{ANC}}}\) | Latent | \(\mathbb{R}^{n}\) | Yes | ||
log_or_gamma |
\(\mathbf{u}_x^{\tilde \gamma}\) | Latent | \(\mathbb{R}^{n}\) |
C.5 NUTS convergence and suitability
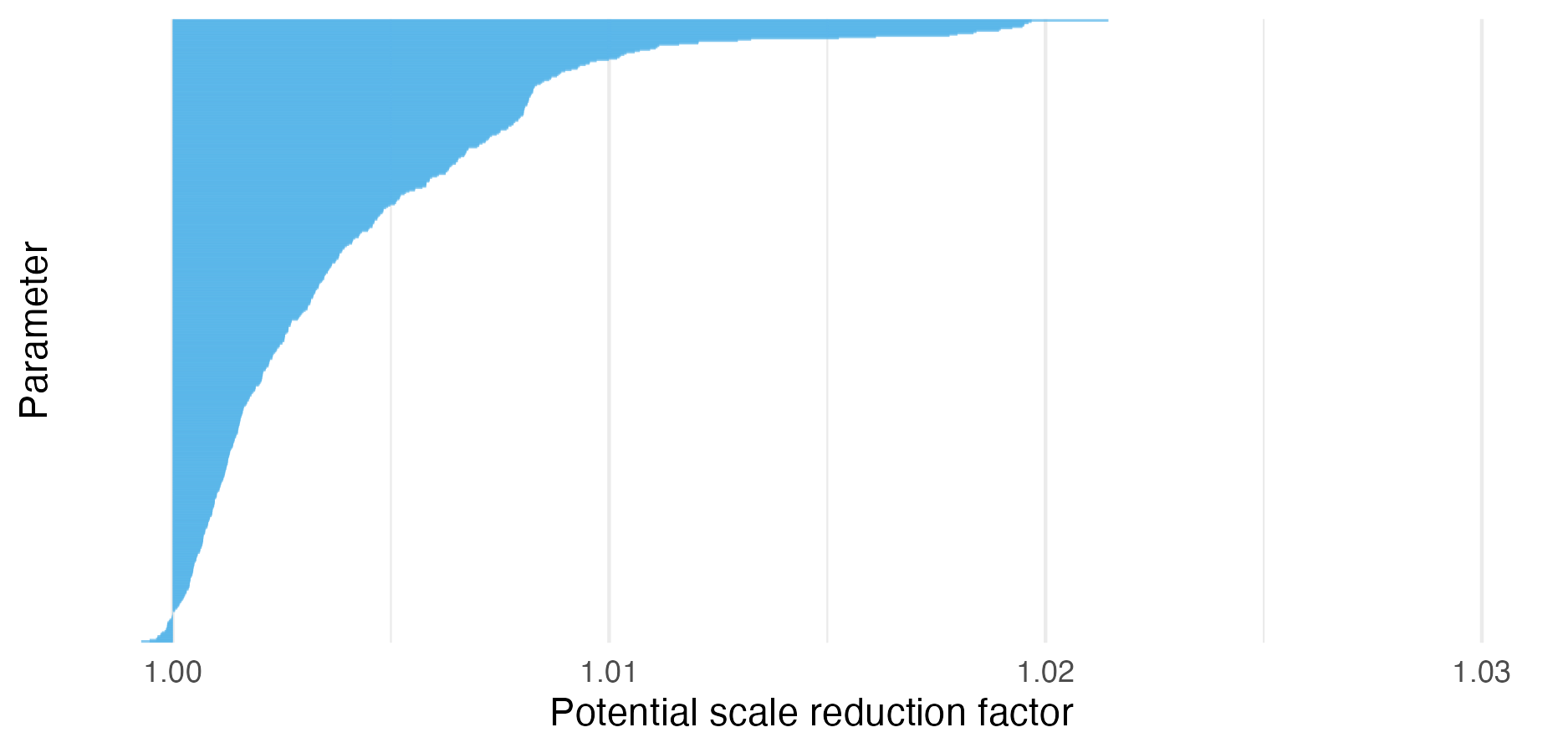
Figure C.5: For NUTS run on the Naomi ELGM, the maximum potential scale reduction factor was 1.021, below the value of 1.05 typically used as a cutoff for acceptable chain mixing, indicating that the results are acceptable to use. Additionally, the vast majority (93.7%) of \(\hat R\) values were less than 1.1.
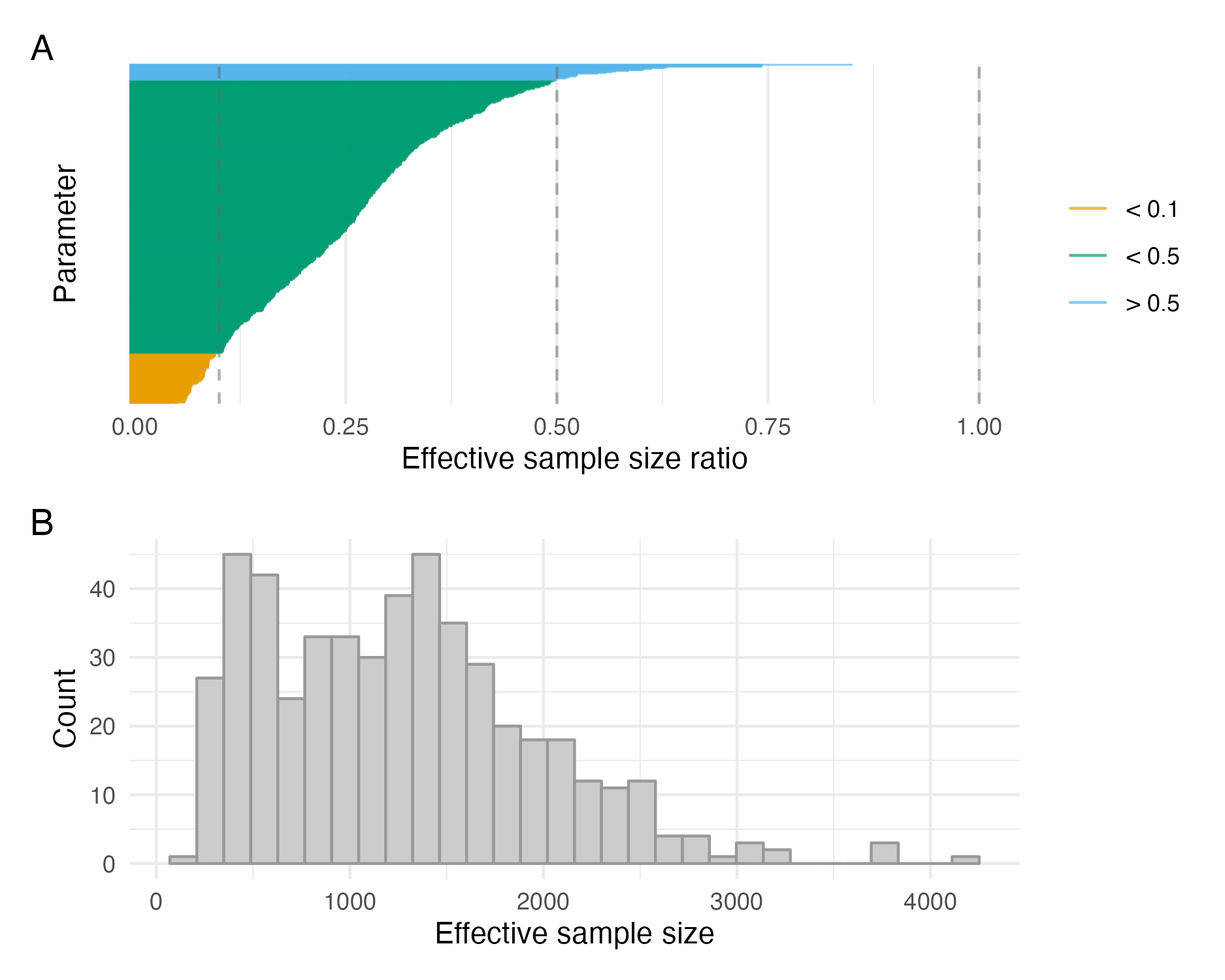
Figure C.6: The efficiency of the NUTS, as measured by the ratio of effective sample size to total number of iterations run, was low for most parameters (Panel A). As a result, the number of iterations required for the the effective number of samples (mean 1265) to be satisfactory was high (Panel B).
![Traceplots for the parameter with the lowest ESS which was log_sigma_alpha_xs (an \(\text{ESS}\) of 208, Panel A) and highest potential scale reduction factor which was ui_lambda_x[10] (an \(\hat R\) of 1.021, Panel B).](resources/naomi-aghq/20231231-180848-af1d67a5/depends/worst-trace.png)
Figure C.7: Traceplots for the parameter with the lowest ESS which was log_sigma_alpha_xs (an \(\text{ESS}\) of 208, Panel A) and highest potential scale reduction factor which was ui_lambda_x[10] (an \(\hat R\) of 1.021, Panel B).
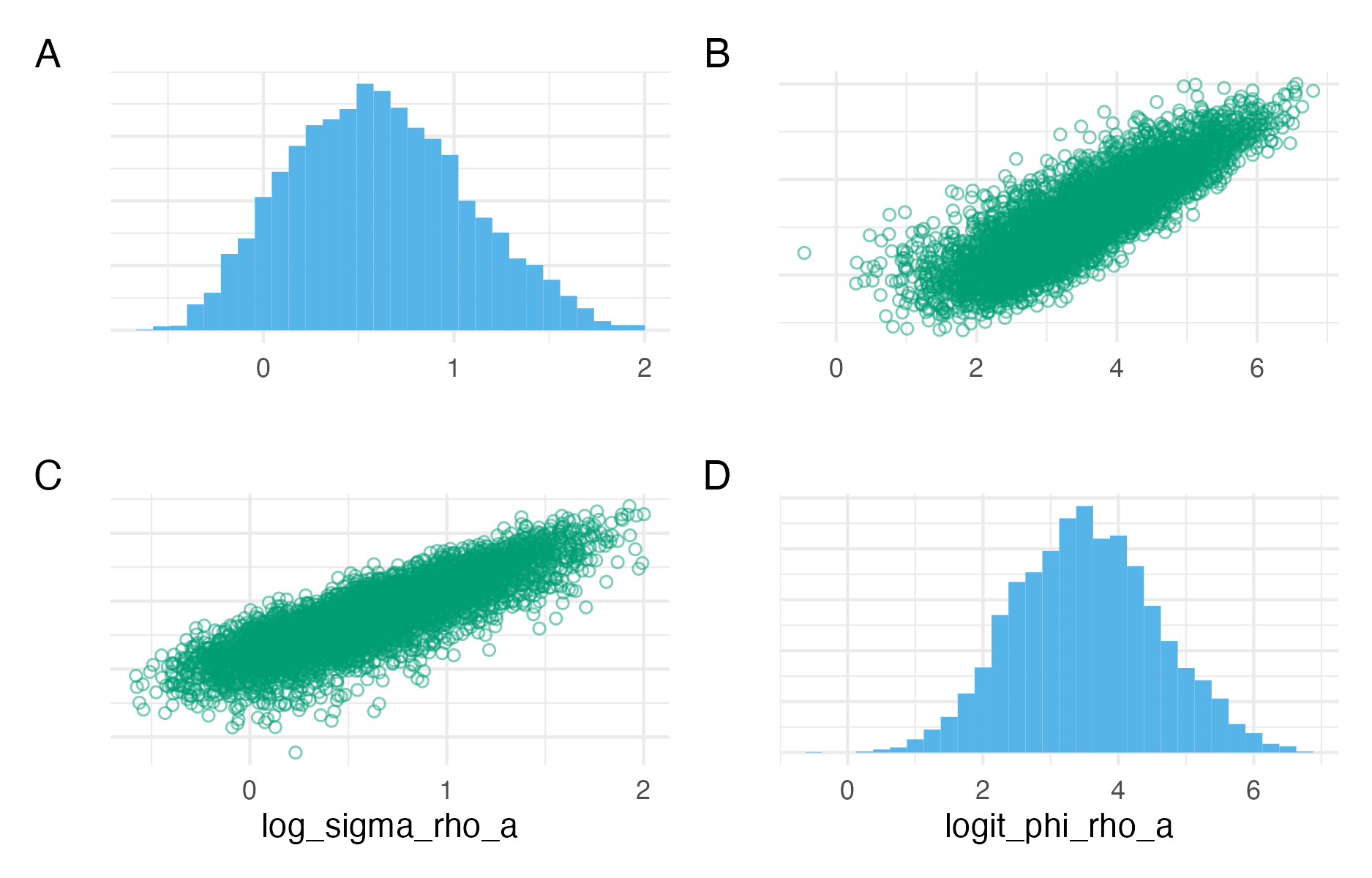
Figure C.8: Pairs plots for the parameters \(\log(\sigma_{A}^\rho)\) and \(\text{logit}(\phi_{A}^\rho)\), or log_sigma_rho_a and logit_phi_rho_a as implemented in code. These parameters are the log standard deviation and logit lag-one correlation parameter of an AR1 process. In the posterior distribution obtained with NUTS, they have a high degree of correlation.
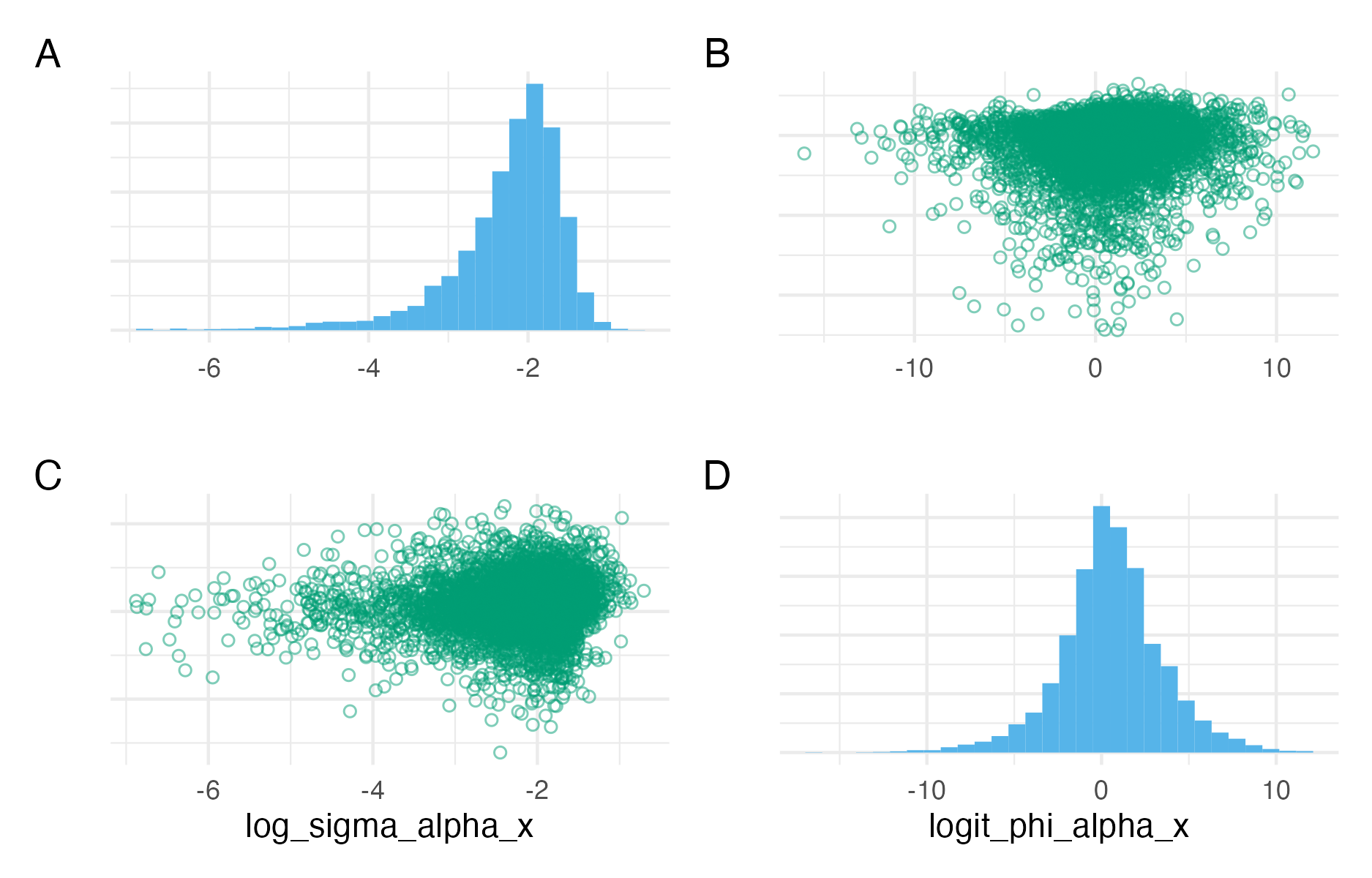
Figure C.9: Pairs plots for the parameters \(\log(\sigma_X^\alpha)\) and \(\text{logit}(\phi_X^\alpha)\), or log_sigma_alpha_x and logit_phi_alpha_x as implemented in code. These parameters are the log standard deviation and logit BYM2 proportion parameter of a BYM2 process. In the posterior distribution obtained with NUTS, they are close to uncorrelated.
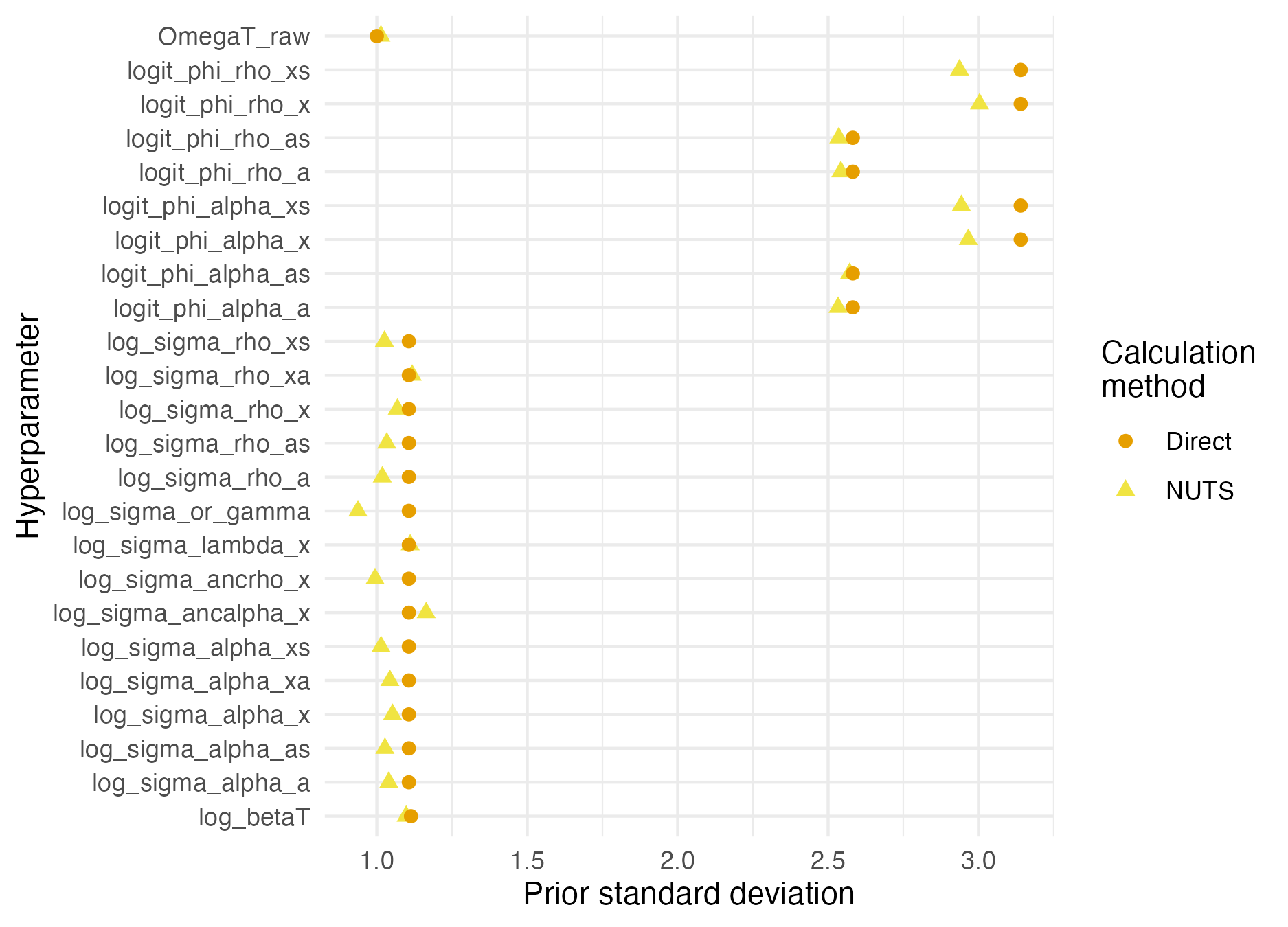
Figure C.10: Prior standard deviations were calculated by using NUTS to simulate from the prior distribution. This approach is more convenient than simulating directly from the model, but can lead to inaccuracies.
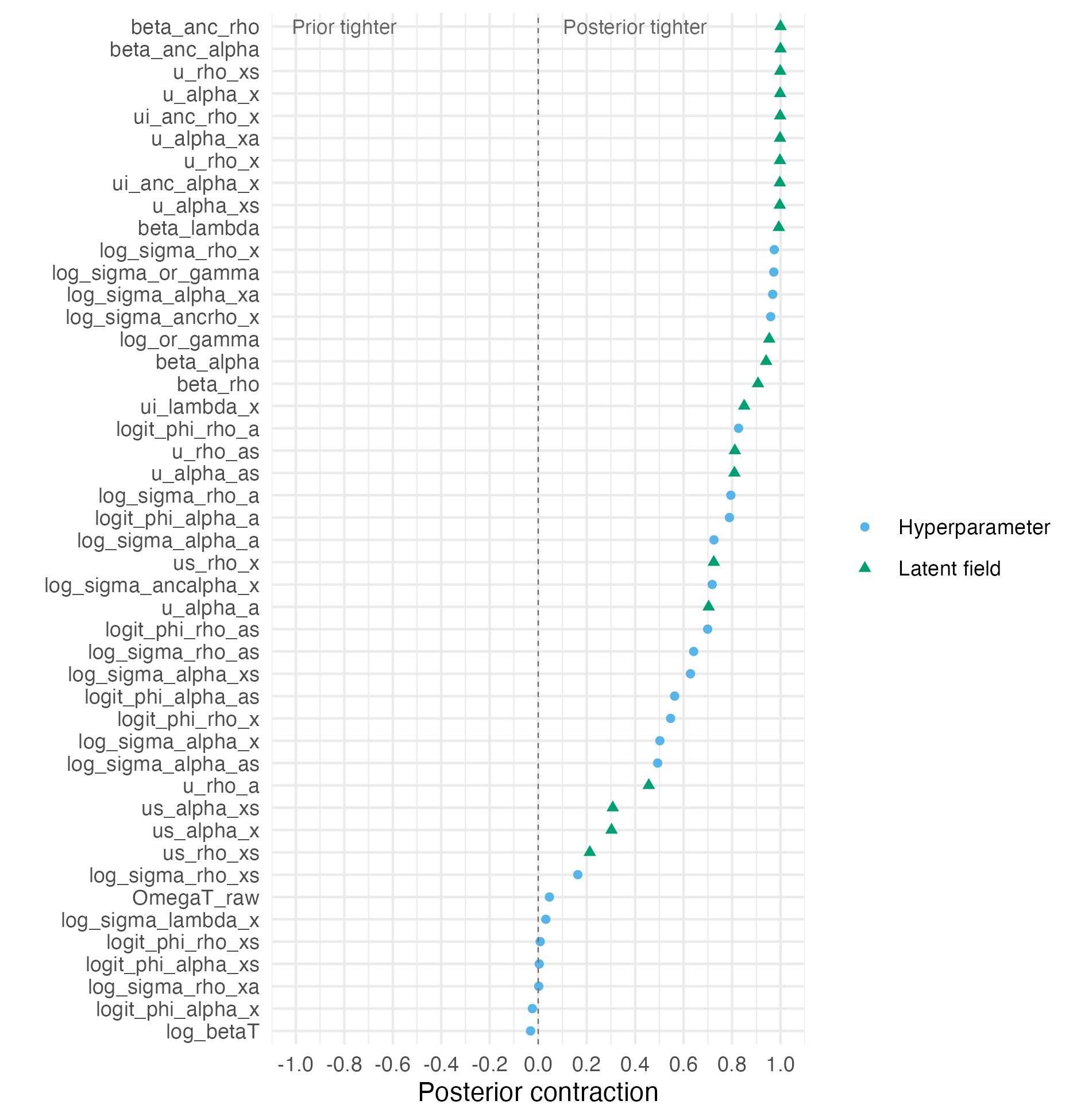
Figure C.11: The posterior contraction for each parameter in the model. Values are averaged for parameters of length greater than one. The posterior contraction is zero when the prior distribution and posterior distribution have the same standard deviation. This could indicate that the data is not informative about the parameter. The closer the posterior contraction is to one, the more than the marginal posterior distribution has concentrated about a single point.
C.6 Use of PCA-AGHQ
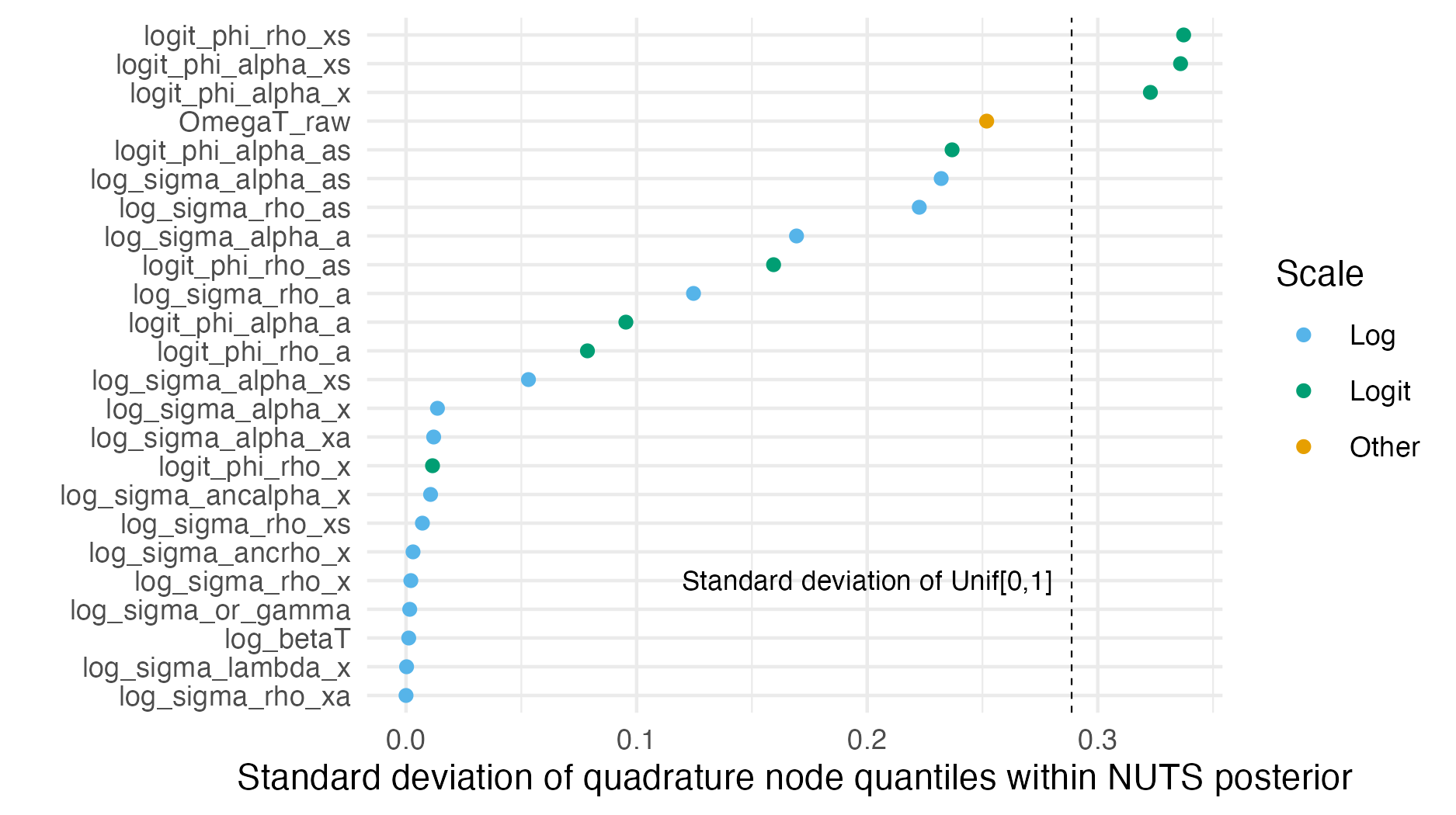
Figure C.12: The standard deviation of the quadrature nodes can be used as a measure of coverage of the posterior marginal distribution. Nodes spaced evenly within the marginal distribution would be expected to uniformly distributed quantile, corresponding to a standard deviation of 0.2867, shown as a dashed line.
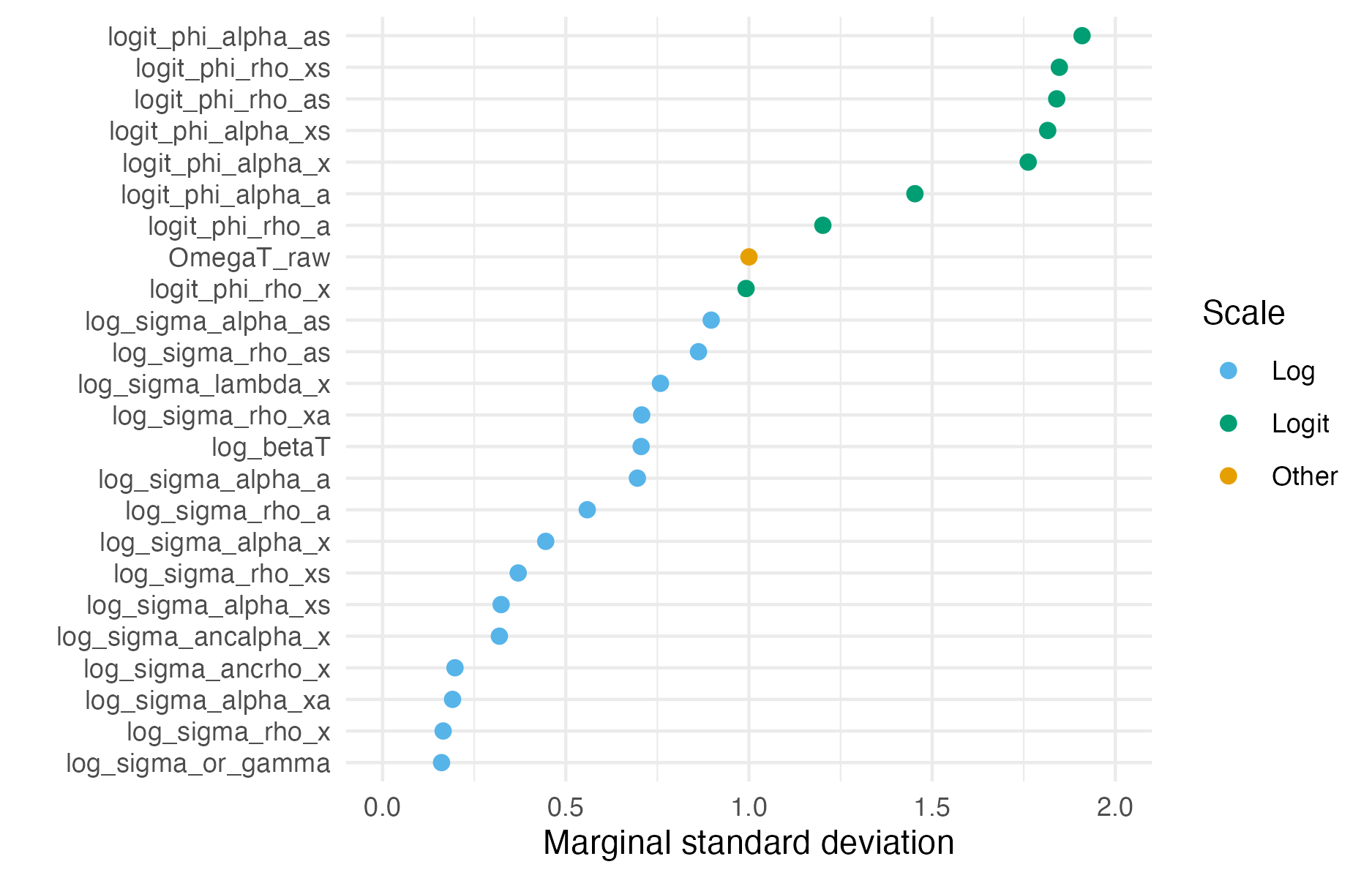
Figure C.13: The estimated posterior marginal standard deviation of each hyperparameter varied substantially based on its scale, either logarithmic or logistic.
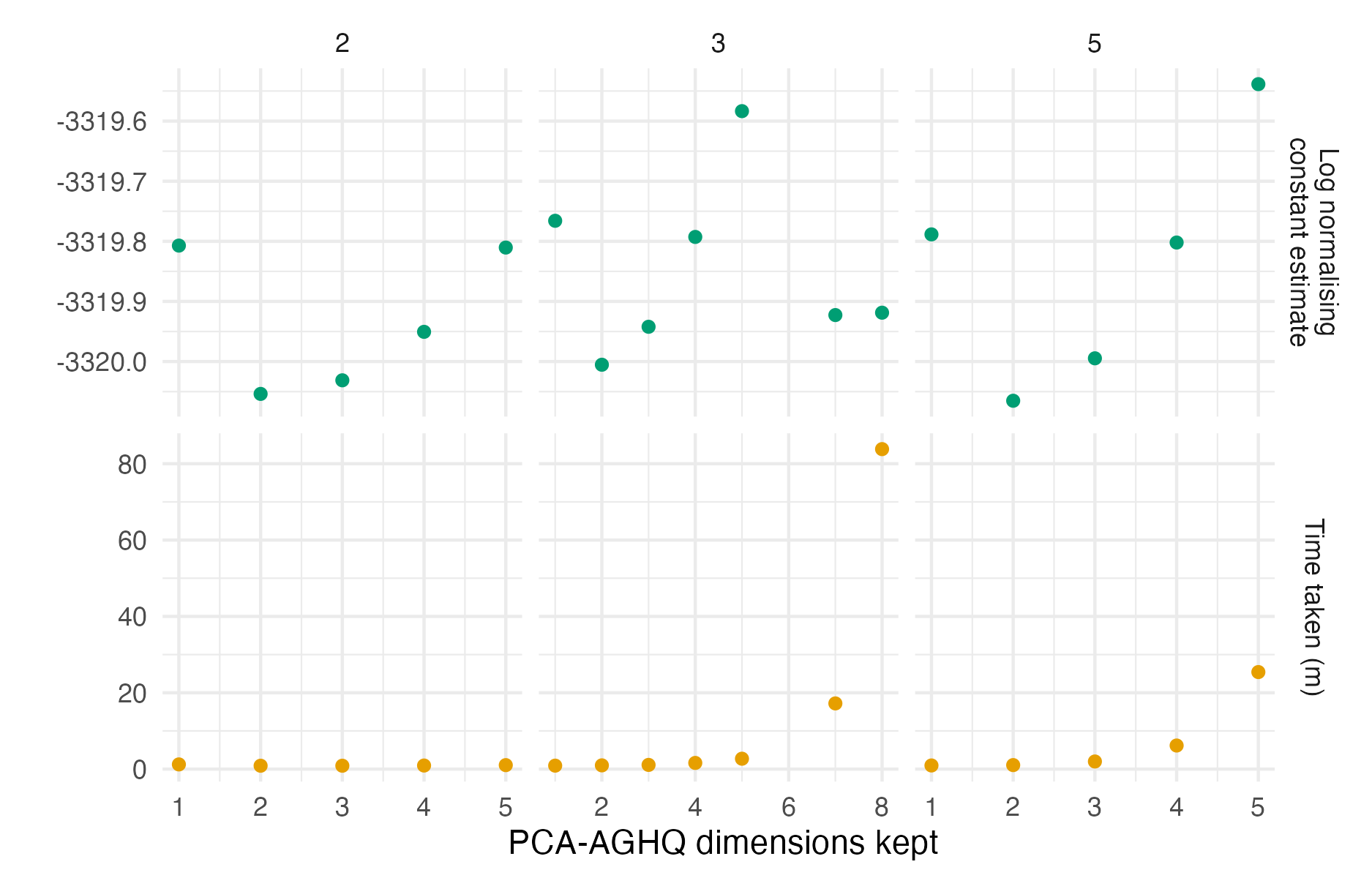
Figure C.14: The logarithm of the normalising constant estimated using PCA-AGHQ and a range of possible values of \(k = 2, 3, 5\) and \(s \leq 8\). Using this range of settings, there was not convergence of the logarithm of the normalising constant estimate. The time taken by GPCA-AGHQ increases exponentially with number of PCA-AGHQ dimensions kept.
C.7 Inference comparison
C.7.1 Point estimates
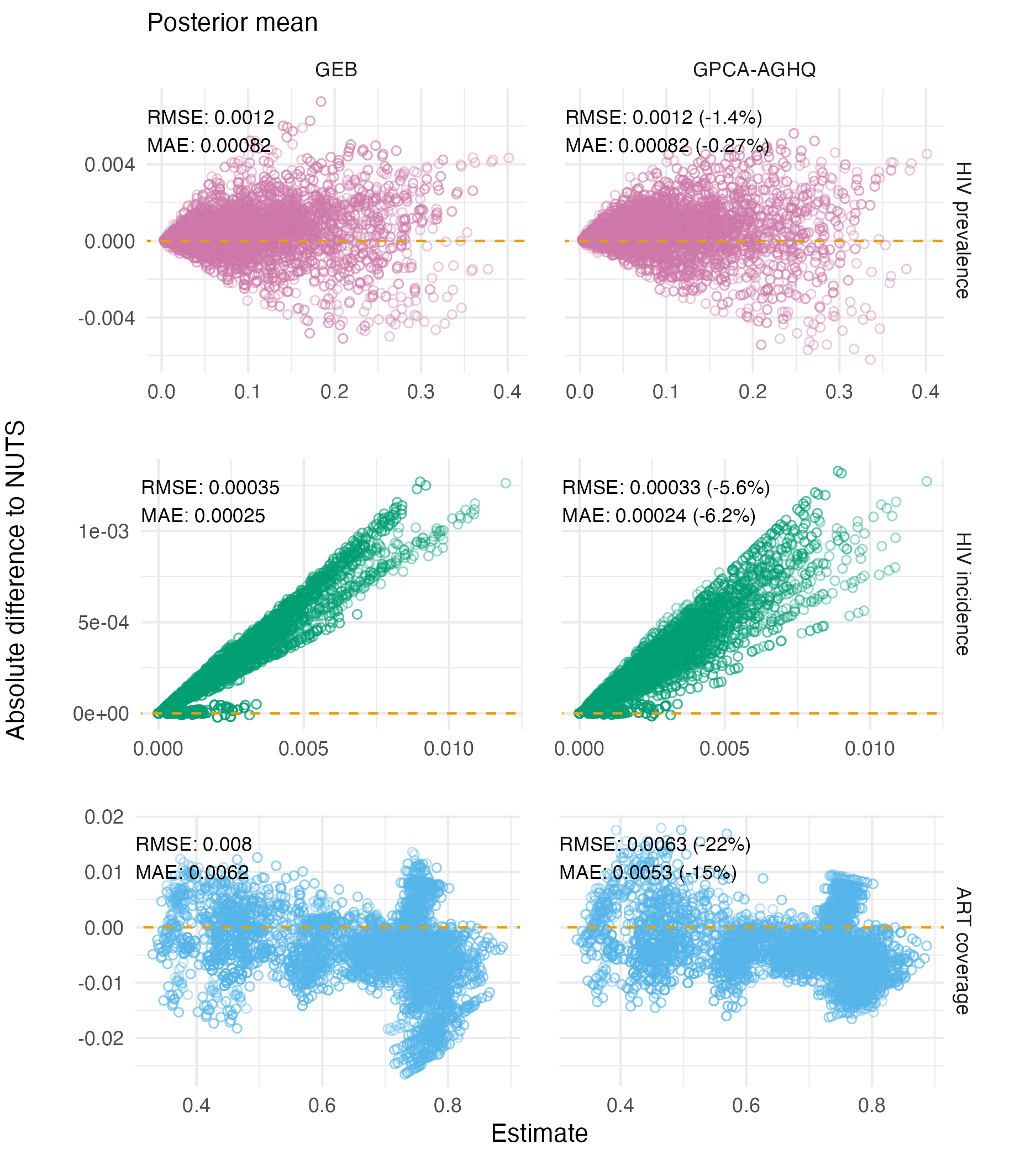
Figure C.15: Differences in Naomi model output posterior means as estimated by GEB and GPCA-AGHQ compared to NUTS. Each point is an estimate of the indicator for a particular strata. In all cases, error is reduced by GPCA-AGHQ, most of all for ART coverage.
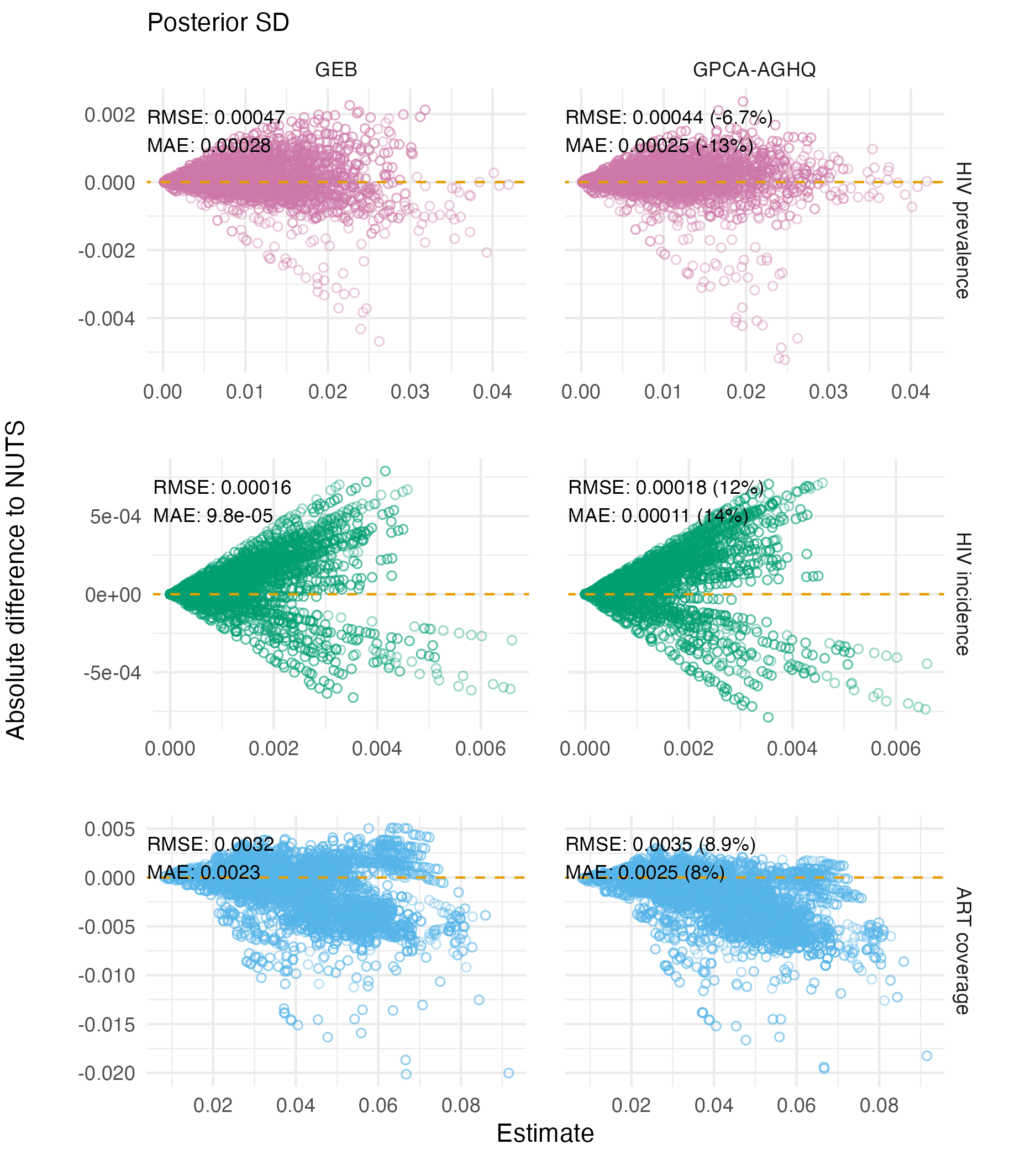
Figure C.16: Differences in Naomi model output posterior standard deviations as estimated by GEB and GPCA-AGHQ compared to NUTS. Each point is an estimate of the indicator for a particular strata. Error is increased by GPCA-AGHQ for HIV prevalence and HIV incidence, and reduced for ART coverage.
C.7.2 Distributional quantities
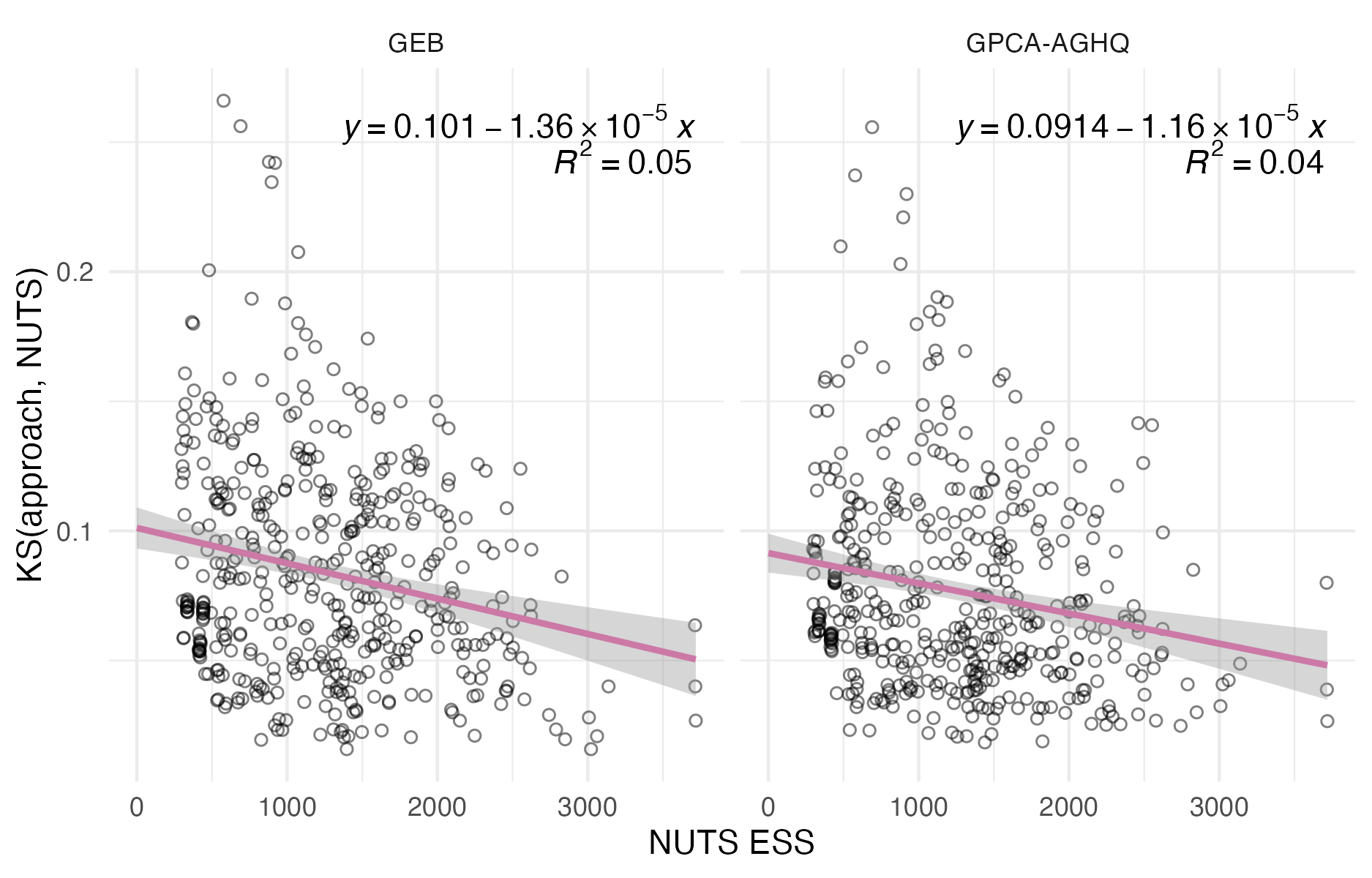
Figure C.17: The Kolmogorov-Smirnov (KS) test statistic for each latent field parameter is correlated with the effective sample size (ESS) from NUTS, for both GEB and GPCA-AGHQ. This may be because parameters which are harder to estimate with INLA-like methods also have posterior distributions which are more difficult to sample from. Alternatively, it may be that high KS values are caused by inaccurate NUTS estimates generated by limited effective samples.